Enterocytozoon hepatopenaei sp. nov. (Microsporida: Enterocytozoonidae)
Enterocytozoon hepatopenaei sp. nov. (Microsporida: Enterocytozoonidae), a parasite of the black tiger shrimp Penaeus monodon (Decapoda: Penaeidae): Fine structure and phylogenetic relationships
Abstract
A new microsporidian species, Enterocytozoon hepatopenaei sp. nov., is described from the hepatopancreas of the black tiger shrimp Penaeus monodon (Crustacea: Decapoda). Different stages of the parasite are described, from early sporogonal plasmodia to mature spores in the cytoplasm of host-cells. The multinucleate sporogonal plasmodia existed in direct contact with the host-cell cytoplasm and contained numerous small blebs at the surface. Binary fission of the plasmodial nuclei occurred during early plasmodial development and numerous pre-sporoblasts were formed within the plasmodium. Electron-dense disks and precursors of the polar tubule developed in the cytoplasm of the plasmodium prior to budding of early sporoblasts from the plasmodial surface. Mature spores were oval, measuring 0.7x1.1microm and contained a single nucleus, 5-6 coils of the polar filament, a posterior vacuole, an anchoring disk attached to the polar filament, and a thick electron-dense wall. The wall was composed of a plasmalemma, an electron-lucent endospore (10nm) and an electron-dense exospore (2nm). DNA primers designed from microsporidian SSU rRNA were used to amplify an 848bp product from the parasite genome (GenBank FJ496356). The sequenced product had 84% identity to the matching region of SSU rRNA from Enterocytozoon bieneusi. Based upon ultrastructural features unique to the family Enterocytozoonidae, cytoplasmic location of the plasmodia and SSU rRNA sequence identity 16% different from E. bieneusi, the parasite was considered to be a new species, E. hepatopenaei, within the genus Enterocytozoon.
1. Introduction
The culture industry for black tiger shrimp Penaeus monodon inThailand has been affected by monodon slow growth syndrome(MSGS) since 2003 (Chayaburakul et al., 2004; Flegel et al.,2004). MSGS-affected shrimp reared in earthen ponds grow at agreatly reduced rate compared to their unaffected counterparts,leading to significant financial losses, comparable to those causedby white-spot syndrome virus (WSSV) outbreaks (Flegel, 2001).The aetiology of MSGS has not been fully elucidated, although mul-tiple infectious agents have been observed in several organs by his-topathology, transmission electron microscopy (TEM), and polymerase chain reaction (PCR) methods (Chayaburakul et al.,2004; Anantasomboon et al., 2006). Most significantly, multiplespheroids were observed in the lymphoid organs and shown tocontain several viruses of known and unknown types (Anantasom-boon et al., 2006; Sritunyalucksana et al., 2006). Hepatopancreaticcells of affected shrimp sometimes also showed the presence ofinclusions of monodon baculovirus (MBV), hepatopancreatic par-vovirus (HPV) or a currently unidentified microsporidian.
Microsporidia are obligate intracellular parasites known to infecta wide range of eukaryotic hosts. Development of the parasite gen-erally occurs within the cytoplasm of the host-cell via nuclear prolif-eration, and spore formation (sporogony), though certain genera areknown to undergo similar development within the host nucleo-plasm (Lom and Dyková, 2002; Stentiford and Bateman, 2007; Sten-tiford et al., 2007). Several genera of microsporidia have beenreported to infect crustacean hosts. These include Agmasoma,Ame-son,Nosema,Pleistophora,Tuzetia,Thelohania,Flabelliforma, Glugoides,Vavraia,Ordospora, Nadelspora and Enterospora (Landg-don, 1991; Larsson et al., 1996, 1997, 1998; Lightner, 1996; Canninget al., 2002; Refardt et al., 2002; Moodie et al., 2003; Amogan et al.,2006; Stentiford and Bateman, 2007). Various crustacean organsand tissues have been reported to be infected (Sprague et al.,1992; Anderson et al., 1989; Landgdon, 1991; Larsson et al., 1996,1997, 1998; Lom et al., 2001). For example, Pasharawipas and Flegel(1994) reported infections of Agmasoma penaei in the muscles andconnective tissue of Penaeus merguensis and P. monodon. Later, Can-ning et al. (2002) reported an infection by Tuzetia weidneri in themuscles of Penaeus setiferus and Penaeus aztecus. Pathogenesis in-volved progression of multifocal lesions to whole muscle blocksand abdominal segments of the shrimp, leading to a conditiontermed ‘cotton tail’ disease. Lightner (1996) has also reported micro-sporidian infections in muscle tissue of penaeid shrimp.
Few microsporidians have been shown to infect only the tubuleepithelial cells of the crustacean hepatopancreas (Anderson et al.,1989; Hudson et al., 2001; Wang and Chen, 2007; Stentiford and Bat-eman, 2007; Stentiford et al., 2007). The publications by Stentifordet al. describe such infections of two European crab species by amicrosporidians placed in a new genus of microsporidia (Enteros-pora) within the family Enterocytozoonidae because of the intranu-clear location of the plasmodia. Placement within the familyEnterocytozoonidae was based upon the formation of precursorsto the spore extrusion apparatus (e.g. polar filament, anchoring disk)and other organelles within the parasite plasmodia (i.e. prior to iso-lation of sporoblasts). This discovery opened the possibility of otherrepresentatives of the Enterocytozoonidae within crustaceans.
The relatively frequent occurrence of a new cytoplamic micro-sporidian that infected only hepatopancreatic tubule epithelialcells of MSGS-affected P. monodon (Chayaburakul et al., 2004) pro-vided an opportunity to study its development and taxonomy. Thisstudy focused mostly on histopathological and ultrastructural fea-tures of the parasite, but also included analysis of a portion of itsSSU rRNA gene and comparison the sequence to those of othermicrosporidians in public databases.
2. Materials and methods
2.1. Samples and investigation process
P. monodon postlarvae (PL) and juveniles were obtained fromhatcheries and commercial ponds experiencing MSGS in Thailand.The age and size of the juveniles varied from one to four months inculture and from 1 to 10 g body weight (BW). The PLs were mostlyPL15 stage, with an approximate BW of 0.1 g. The hepatopancreaswas dissected out and portions were smeared onto glass micro-scope slides and stained with haematoxylin and eosin (H&E). Otherhepatopancreatic tissue specimens fixed, embedded, sectioned andstained for light (LM) and transmission electron (TEM) microscopy.Samples from individuals positively diagnosed with microsporidi-an infections via smears or histology were processed for ultrastruc-tural investigation. The size of the microsporidian spores wasdetermined from fresh spores prepared by differential centrifuga-tion of homogenized, infected hepatopancreatic tissue ending withPercoll (GE Life Sciences) density gradient centrifugation. Thespore band was removed from the gradient and examined directlyin wet mounts. For LM and TEM, 10 juveniles and 10 PL15 weresampled. For molecular biology, DNA was extracted from morethan 1000 juveniles and 10,000 PL15.
2.2. Histology and electron microscopy
For LM, a mid-sagittal cut was performed on infected shrimpwhich were then fixed in Davidsons’ seawater fixative for 24 h, processed routinely for histology, sectioned at 5lm and stainedwith haemotoxylin and eosin (H&E), Giemsa, and Chromotrope2R modified trichrome (Weber et al., 1992). For TEM, small pieces(2 mm3) of hepatopancreatic tissue from the same shrimp speci-mens were fixed in 2.5% glutaraldehyde and 4% paraformaldehyde,or 6% glutaraldehyde, in Millonig buffer followed by washing inthree changes of 0.1 M sodium cacodylate buffer and stained enbloc in 0.5% aqueous uranyl acetate for 1 h. The fixed tissues wereembedded in epoxy resin 812 (Agar Scientific-pre-mix kit 812) fol-lowing dehydration through a graded acetone series. Thick sectionswere stained with Toluidine Blue for viewing with a light micro-scope to identify sections with suitable target areas. Ultra thin sec-tions (70–90 nm) of these samples were mounted on uncoatedcopper grids and stained with uranyl acetate and Reynolds’ leadcitrate. Sections were examined using a JEOL 1210 transmissionelectron microscope.
2.3. DNA extraction, PCR and sequencing
Total DNA was extracted from the hepatopancreas using a com-mercial tissue extraction kit (Qiagen, Germany). DNA was ampli-fied using PCR primers designed from the alignment of 11microsporidian SSU rRNA gene sequences at GenBank in such amanner that the primers were based in highly conserved regionsof the gene but amplified a fragment from a highly variable region.The primers were MF1 (forward) 50-CCG GAG AGG GAG CCT GAGA-30and MR1 (reverse) 50-GAC GGG CGG TGT GTA CAA A-30, rela-tive to positions 242–260 and 1165–1183, respectively, of thesmall subunit (SSU) rRNA gene of Enterocytozoon bieneusi (Genbankaccession No. AF024657). Based on the 11 sequences used, ex-pected amplicons would be in the range of approximately 900–1000 bp. The PCR process was carried out in 50lm reaction mix-tures containing PCR buffer, 200 mM dNTP, 2 mM MgCl2, 1.25 unitsTaq polymerase, 1 mM primers and 1ll template. Reactions werefollowed by 35 cycles of denaturation for 30 s at 94 °C, annealingfor 30 s at 55 °C, and extension at 72 °C for 90 s, followed by a 5-min final extension at 72 °C. An SSU fragment of 1200 bp in lengthwas amplified and inserted into a TOPO Cloning Kit (Invitrogen,USA). Plasmids were checked for inclusion of the desired insertand subsequently purified with NucleoSpin Extract Kit (Qiagen,Germany). Following hybridization with shrimp genomic DNA,clones that did not show signals were further processed for DNAsequence determination (Macrogen, South Korea). The insert fromone of these clones (final length 848 bp minus the primer se-quences) gave high sequence identity to microsporidian rRNA se-quences in the GenBank database using a BLASTn (http://www.ncbi.nlm.nih.gov/BLAST) search and was sequenced fromboth strands. It was aligned with sequences of other microsporid-ians in the database using CLUSTAL W 1.7 multiple alignment soft-ware (Thompson et al., 1994). The phylogenic tree was constructedusing CLUSTAL W 1.75 program with the microsporidian Amblyos-pora canadensis as an out-group since Amblyospora is from Clade 1of the microsporidians (Vossbrinck and Debrunner-Vossbrinck,2005). Tree topology was evaluated by bootstrap analysis usingthe neighbour joining method with the program parameter setfor 1000 replicates.
3. Results
3.1. Histopathology and ultrastructure
Hepatopancreatic samples prepared by the smear method andviewed by light microscopy (LM) showed the presence of micro-sporidian spores (Fig. 1A) only in the cytoplasm of hepatopancreat-ic tubule epithelial cells. Using fresh spore preparations from Percoll gradient separations (Fig. 1B), spore size was1.1 ± 0.2lm0.7 ± 0.1lm(N= 100), while that in HP smearswas 1.1 ± 0.2lm0.6 ± 0.2lm(N= 10). Histologically, spores ap-peared as acidophilic structures confined within a vacuole withinthe cytoplasm of R (reserve), B (blister) and E (embryonic) cells,but F (fibrillar) cells (Fig. 1C and D). However, the midgut and hind-gut epithelial cells, or cells of other tissues and organs were unaf-fected. Histological sections revealed several developmental stagesof the microsporidian, depicted by differences in the staining ofvarious cytoplasmic inclusions (Fig. 1E). Mature spores appearedto contain electron-lucent regions that likely coincided with theposterior vacuole observed by TEM (see below).
TEM revealed several stages of a microsporidian parasite inthe cytoplasm of hepatopancreatic, tubule epithelial cells. Defin-itive early meront stages were not observed but early and latestage plasmodia were frequent and showed multiple nuclei andsporogony typical of members of the family Enterocytozoonidae(Fig. 2A and B). The plasma membrane of some plasmodia was in direct contact with the host-cell cytoplasm (Fig. 2A and B),whereas some were apparently surrounded by cytoplasmic vacu-oles, often encompassing multiple, discrete plasmodia (Fig. 2C).Plasmodia were delimited by an electron dense membrane thatseparated plasmodial components from the host-cell cytoplasm(Fig. 2A and B). Evidence of nuclear division, or binary fissionwas observed in some plasmodia, characterized by the presenceof spindle plaques on the outer surface of the nuclear envelope(Fig. 2C, inset).
Sporogonal plasmodia were characterized by the appearance ofround vesicles (100–200 nm) with electron-dense, surroundingmembranes and short polar filament precursors within the plas-modial cytoplasm (Fig. 3A, inset a). Circular disks with outer elec-tron dense layers and inner electron-lucent cores were alsoobserved (Fig. 3A, inset b). The tubules and the circular disks werepresumably precursors of the polar filaments of the spore. As plas-modia matured, the tubules and the disks appeared to increase innumber, concomitant with a decrease in the number of vesicles(Fig. 3B). Polar filament precursors increased in length and in cur-vature to form several distinct coils. At this stage, the parasite nu-clei were oriented toward the periphery of the plasmodium, withpolar filament coils located on the inner side (Fig. 3C).
The sporogonal, plasmodial membrane, which was in directcontact with the host-cell cytoplasm, displayed a microvillousappearance with characteristic surface blebs (Fig. 3D and E). Themembrane on which these blebs occurred was externally rein-forced by a dense coat and was composed of two inner membra-nous layers and an electron-dense outer layer of 10 nmthickness. The membrane of the microvillous structure was alsothickened, but it was somewhat thinner than that in the regionof the blebs. Surrounding the plasmodium was host-cell cytoplasm,with abundant rough and smooth endoplasmic reticula (RER andSER), clear vesicles and mitochondria.
As development progressed, pre-sporoblast units (containing asingle nucleus and elements of the spore extrusion apparatus)were located towards the periphery of the plasmodium, with upto 20 pre-sporoblasts visible within a single plasmodial section(Fig. 3F). A space was formed between the coils of the polar fila-ment and the nucleus, which was occupied by the early posteriorvacuole (Fig. 4A). A dense cap, a precursor of the anchoring disk,was also formed at the anterior end of the pre-sporoblast. The pos-terior vacuole apparently became larger as the sporoblast matured(Fig. 4B). The coiled polar filaments (5–6 coils) also appeared to in-crease in diameter to approximately 75 nm with the innermostelectron-dense core surrounded by an electron-lucent layer andan outer electron-dense coat. A double-layered nuclear membranewas clearly observed at this stage.
Pre-sporoblasts protruded to form buds from the plasmodialmembrane that apparently separated to form electron-densesporoblasts that lay in direct contact with the host cytoplasm(Fig. 5A). Sporoblasts underwent direct development to mature,electron dense and oval spores. The spores contained a single nu-cleus, with a polar filament coiled 5–6 times at the posterior endand terminating at an anchoring disk at the anterior end(Fig. 5B–D). The posterior vacuole was occasionally visible withinmature spores. The wall of the spore was composed of a cell mem-brane within an electron-lucent endospore (10 nm) and an elec-tron-dense exospore (2 nm).
3.2. SSU rRNA gene fragment analysis
PCR using primers based on conserved regions of microsporidi-an SSU rRNA sequences listed at GenBank and DNA template frommicrosporidian-infected P. monodon hepatopancreatic tissueyielded an amplicon of 886 bp which was in the expected ampliconrange of approximately 900–1000 bp. After cloning and sequenc-ing, a fragment of 848 bp (excluding the primer sequences) wassubjected to a general BLASTn search that yielded hits only formicrosporidian sequence records. Top hits included Nucleosporasalmonis (AF185998) at 87% identity and E. bieneusi (GenBankAF023245) at 86% identity. By contrast, sequence identity for A. pe-naei (the causative agent of ‘cotton shrimp’ reported from P. mon-odon in Thailand) was only 71%. In addition, the sequence identitymatch between N. salmonis and E. bieneusi (from different genera inthe family Enterocytozoonidae) was 85%. These results indicatedthat the 848 bp sequence from P. monodon was novel and approx-imately equidistant from N. salmonis and E. bieneusi in terms ofidentity difference. A subsequent CLUSTAL W alignment was car-ried out and a phylogenetic tree was constructed with the optimalcriteria set for distance and using SSU rRNA gene sequences ofavailable microsporidians in the public database. The un-rootedtree constructed using this data (Fig. 6) revealed that the sequenceof the microsporidian from P. monodon most closely grouped withN. salmonis and E. bieneusi.
4. Discussion
This is the first report on ultrastructural features and a partialSSU rRNA gene sequence of a microsporidian exclusively infectinghepatopancreatic tubule epithelial cells of the black tiger shrimp P.monodon. Its unique ultrastructural features linked it to the familyEnterocytozoonidae. Based on its distinction from the microsporid-ian genera Nucleospora and Enterospora in the family and on itssimilarity to the single species in the genus Enterocytozoon, we as-signed the new microsporidian infecting P. monodon to the genusEnterocytozoon. We believe that its occurrence in an invertebratehost and its relatively low SSU rRNA sequence identity with E. bien-eusi justify its assignment to a new species within the genus.
The name E. hepatopenaei is proposed for the new species withjustification based on ultrastructural characters that conform tothe unique features of the family Enterocytozoonidae but distin-guish it from other species in the family, i.e., E. beiniusi that alsodevelops in the host-cell cytoplasm and species in the genera Enterospora and Nucleospora that develop within the host-cell nucleus.Distinction from Enterospora and Nucleospora is clear based theirintranuclear rather than cytoplasmic development. However, dis-tinction from E. bieniusi is not initially so clear.
E. hepatopenaei has several features similar to E. bieneusi, theonly current member in the genus Enterocytozoon (Franzen andMüller, 1999). For both species, the sporogonal plasmodium islocalized in the host-cell cytoplasm and it generates pre-cursorsof the spore extrusion apparatus prior to the formation and bud-ding of sporoblasts. For both, sporoblasts are liberated to the hostcytoplasm following which they develop directly into oval spores.Other similar morphological features included spores with a pos-terior vacuole and 5–7 coils of polar filaments arranged in tworows (Franzen and Müller, 1999; Wasson and Peper, 2000; Vis-vesvara, 2002; Didier, 2005). However, the mean spore size of E.hepatopenaei (1.1 0.7lm in fresh preparations) was somewhatsmaller than that of E. bieneusi (1.5 0.5–1.0lm) (Franzen andMüller, 1999; Didier et al., 2000; Wasson and Peper, 2000),although the difference was not great, and it might be argued thatit was not sufficient to warrant designation of two separatespecies.
Since only monokaryotic nuclei have been reported from infec-tions of E. bieneusi in vertebrates, it is conceivable that other lifecycle stages such as diplokaryotic forms and gametes might occurin an alternative host, and that shrimp could constitute such a host.However, the fact that the spores in P. mondon are also monokary-otic and that they develop in a manner similar to those of E. bien-eusi rather than developing from diplokaryotic or meiotic nuclei(Flegel and Pasharawipas, 1995) argues against this possibility. Inaddition, work on possible sources of E. bieneusi in human infec-tions has implicated birds and mammals rather than shrimp, andfresh water sources rather than marine (Dowd et al., 1998; Didieret al., 2004; Lobo et al., 2006). All this information argues in favorof our contention that E. hepatopenaei is distinct from E. bieneusi.
The argument for distinction of the two species is further sup-ported by the 16% difference in identity for compared fragmentsof their SSU rRNA genes. A rough estimation of the sequence iden-tity cutoff for a species is approximately 97% (Stackebrandt andGoebel, 1994) so that 84% identity is well outside the range of whatwould be considered reasonable for con-specific isolates. Indeed,the fact that N. salmonis and E. bieneusi are in different generaand differ by 85%, might suggest that the hepatopancreatic micro-sporidian in P. monodon should also be placed in a new genus.Thus, our proposal for a new species within the genus Enterocyto-zoon rather than a new genus may be viewed as conservative.
With respect to the phylogenetic tree, it may be argued thatcomparison using 848 bp of a single gene does not cover a suffi-cient proportion of the target genomes to prepare a robust tree. De-spite this caveat, the relationships among the databasemicrosporidians in our phylogenetic tree (Fig. 6) are similar tothose reported for them in more comprehensive comparisons (Vos-sbrinck and Debrunner-Vossbrinck, 2005). In any case, our preli-minary analysis based solely on this region indicates that E.hepatopenaei tends to group with other microsporidians associatedwith aquatic host species. This intriguing relationship may be fol-lowed up using longer sequences and more genes whenever thisinformation becomes available.
For other members of the family Enterocytozoonidae previouslydescribed from crab hosts (i.e., genus Enterospora, Stentiford, 2007)there is no available genetic information. However, they differfrom E. hepatopenaei greatly in that they develop within the hostnucleus rather than the cytoplasm. In addition, E. hepatopenaei in-fected all cell types of the hepatopancreatic tubule epithelium ex-cept F-cells while Enterospora canceri infected all cell types exceptB-cells (Stentiford et al., 2007; Stentiford and Bateman, 2007).
The only other microsporidian described from penaeid shrimpin Thailand is Agmasoma penaei (Pasharawipas and Flegel, 1994)that causes infections in muscle and connective tissue. The nucleiin the plasmodia are diplokaryotic and give rise to monokaryoticnuclei by reductive sporogony (Flegel and Pasharawipas, 1995).Further, the SSU rRNA sequence of A. penaei (Pasharawipas et al.,1994; GenBank) shared only 71% identity with that of E. hepatope-naei reported herein. They are obviously only distantly related.
Several microsporidian species develop directly within thehost-cell cytoplasm and not within a vacuole. However, Encephali-tozoon spp. and Glugoides intestinalis possess a parasitophorousvacuole at an early stage of infection that probably acts as a protec-tion against destruction within the host-cell cytoplasm (Cali andKolter, 1993; Cali and Takvorian, 1999; Vavra and Larsson, 1999).In E. hepatopenei, it was observed that some plasmodia were con-tained within a vacuole while some were not. However, blebsand microvilli-like structures were present in the thick membranesurrounding plasmodia that were not contained in vacuoles. It ispossible that plasmodia begin as small structures in vacuoles andthen expand until regions of the plasmodial membrane come intodirect contact with the vacuolar membrane to form what appearsto be a thick, double-layered membrane. If so, lack of complete opposition of the membranes could lead to the observed blebs andmicrovillus-like structures. A similar thin, electron-dense amor-phous layer on the external surface of the plasmodial membranehas also been observed in the microsporidian Vavraia mediterranicainfecting the muscle of the shrimp Crangon crangon (Azevedo,2001). Unfortunately, no rRNA gene sequence information is avail-able for this species.
Abundant RER and mitochondria were found in the host cyto-plasm surrounding E. hepatopenaei, but not within the cytoplasmof plasmodia. This is normal for microsporidia. Many now believethem to constitute a Phylum in the fungal kingdom and torepresent reduced forms that have lost their mitochondria andother organelles through adaptation to their intracellular para-sitic life-style (Lee et al., 2008; Keeling et al., 2005; Keelingand Fast, 2002; Fischer and Palmer, 2005; James et al., 2006;Hibbett et al., 2007). This is supported not only by genesequence homology but also by similarity in steps of the meioticdivision cycle and in possession of paired nuclei in diplokaryons(microsporidia) and dikaryons (fungi) (Flegel and Pasharawipas,1995).
The hepatopancreatic microsporidian infections we studied oc-curred most frequently in MSGS P. monodon that were concomi-tantly infected with several viruses (Chayaburakul et al., 2004)orwith pathogens in the lymphoid organ (Anantasomboon et al.,2006) or hematopoietic tissue. They were less frequent in normalshrimp. Thus, it is possible that the microsporidian infections wereopportunistic in nature, with the pathogen exploiting a weakenedimmune status of the host. For example, the occurrence of humanand mammalian infections of E. bieneusi in enterocytes of the duo-denum and ileum of AIDS patients is dependent on chronic immu-nosuppression (Desportes et al., 1985).
5. Taxonomic summary
Phylum: Microspora (Sprague, 1977).
Class: Microsporea (Delphy, 1963).
Order: Microsporida (Balbiani, 1882).
Family: Enterocytozoonidae (Cali and Owen, 1990).
Genus: Enterocytozoon (Desportes et al., 1985).
5.1. E. hepatopenaei n. sp.
Specific diagnosis: Spores ovoid, measuring 0.7 1.1lm in freshpreparation (shorter than those of E. bienneusi) and containing 5–6visible coils of the polar filament. Polar filament precursors andother spore organelles formed within the sporogonal plasmodiumand packaged into pre-sporoblast units prior to budding of sporo-blasts to the host-cell cytoplasm.
Type host: P. monodon L. (Decapoda: Penaeidae)
Type locality: Chantoburi Province, eastern coast of Thailand
Site of infection: Cytoplasm of the tubule epithelial cells of thehepatopancreas
Etymology: The specific epithet relates to the location of theinfection in hepatopancreatic tissue and to the generic name (Pena-eus) of the host shrimp, thus hepatopenaei.
Type material: Paraffin and plastic blocks containing infectedmaterial have been deposited in the Center of Excellence forShrimp Molecular and Biotechnology (Centex shrimp), MahidolUniversity, Thailand.
Gene sequence: The partial SSU rDNA sequence of the microspo-ridian, E. hepatopenaei, has been deposited in the GenBank/EMBLdatabase under Accession No. FJ496356.
Acknowledgments
We would like to thank Dr. Jessada Denduangboripant and Ms.Linda Nelson for their help in phylogenetic tree analysis. We wouldalso like to thank Dr. T.W. Flegel for assistance in editing the man-uscript. This work was supported by a Mahidol University ResearchGrant 2002–2006.

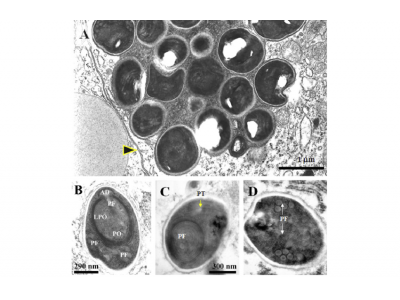
Fig. 1. Photomicrographs of Enterocytozoon hepatopenaei tubule epithelial cells of the hepatopancreas of Penaeus monodon. (A) H&E-stained smear of hepatopancreatic tissue showing numerous microsporidian spores (arrows). (B) Fresh preparation of microsporidian spores from a Percoll gradient. (C and D) Hepatopancreatic tissue sections showing acidophilic, granular inclusions in the cytoplasm of tubule epithelial cells (arrows). (E) Semi-thin section of hepatopancreatic tissue showing early and late plasmodia (inset a) and mature spores (inset b) in the cytoplasm of tubule epithelial cells. Some spores show unstained spots that represent their concave surfaces at one end (inset b). A, haematoxylin stain; C, Trichrome stain; D, H&E stain; E, Toluidine blue stain. ePm, early plasmodium; lPm, late plasmodium.

Fig. 2. Trasmission electron micrographs (TEM) of early stages of E. hepatopenaei development in hepatopancreatic tubule epithelial cells of P. monodon. (A) Early plasmodium stage with two nuclei. (B) Early plasmodium stage with multiple nuclei. (C) A group of plasmodia surrounded by a vacuole within the cytoplasm of the host-cell and showing nuclear division (inset, arrowhead). Mv, microvilli; V, vacuole.

Fig. 3. TEM of sporogonial stages of E. hepatopenaei development in hepatopancreatic tubule epithelial cells of P. monodon. (A) Early sporogonial plasmodium containing nuclei (asterisks), clear vesicles (inset a, arrowhead), and precursors of polar filaments (inset a, arrows; and inset b). (B) Sporogonial plasmodium with peripherally located sporoplasm (asterisk). (C) Sporogonial plasmodium with several peripherally located pre-sporoblasts (asterisks). (D) Plasmodial membrane showing a microvillus-like structure surrounded by host-cell cytoplasm (arrow) containing extensive RER (arrowhead). (E) Thickened plasmodial plasmalemma showing vesicular structures at the interface with host cytoplasm (arrowhead). (F) Late sporogonial plasmodium showing assembled pre-sporoblasts, polar filaments and a dense cap (arrowhead).

Fig. 4. TEM of E. hepatopenaei sporoblasts in hepatopancreatic tubule epithelial cells of P. monodon. (A) Sporoblast at the periphery of a sporogonal plasmodium showing the nucleus, early posterior vacuole (asterisks), immature polar filaments with five cross sections (arrowhead) and a dense cap or anchoring disk (arrow). (B) Late stage sporoblast showing mature polar filaments (arrowhead). N, nucleus; PV, posterior vacuole.

Fig. 5. TEM of E. hepatopenaei spore development. (A) Immature spores covered with the plasmodial membrane (arrow). (B) Immature spore showing the anchoring disk (AD), the polar filament (PF) and the polaroplast (PO) including its lamellar portion (LPO). (C) Cross section of a mature spore just below the anchoring disk showing a cross section of the polar filament (PF) surrounded by the lamellar portion of the polaroplast (LPO) and a longitudinal section of the polar filament outside the polaroplast. (D) Mature spore showing a section of the coiled portion of the polar filament. The scale bar on (c) is common for (d).

Fig. 6. Phylogenetic tree of E. hepatopenaei and selected microsporidians derived by using a CLUSTAL W alignment of matched SSU rRNA gene sequences with the optimalitycriterion set for distance and with Amblyospora canadensis as an out-group. Numbers at branch points indicate bootstrap values for 1000 replicates
Authors:
Somjintana Tourtip, Somja Wongtripop, Grant D. Stentiford, Kelly S. Bateman, Siriporn Sriurairatana, Jittipan Chavadej, Kallaya Sritunyalucksana, Boonsirm Withyachumnarnkul
Related news
 A comprehensive look at the Proficiency Test for farmed shrimp
A comprehensive look at the Proficiency Test for farmed shrimp Based on the importance of infectious diseases in the shrimp industry worldwide and the problems associated with their presence, the diagnosis of shrimp pathoge
 Asepsis key to prevent contamination in shrimp hatcheries
Asepsis key to prevent contamination in shrimp hatcheries Maintaining biosecurity in shrimp hatcheries by minimizing risk factors and the presence of pathogens and their potential dispersion is key to the success
 Organic acids in aquafeeds: A potential substitute for antibiotics
Organic acids in aquafeeds: A potential substitute for antibiotics Using the right types can enhance growth, nutrient utilization, immune response and disease resistance
 Shrimp trial sets stage for insect protein in aquafeed
Shrimp trial sets stage for insect protein in aquafeed The white leg shrimp is one of the main species in aquaculture. 3.7 million metric tons are produced every year
 Mixed feeding schedules benefit growth of Pacific shrimp
Mixed feeding schedules benefit growth of Pacific shrimp Aquafeeds are the most important cost in fed fish and shrimp aquaculture operations, especially considering that – even under appropriate feeding management